Phytoremediation: Difference between revisions
m Corrected "ox" to "of" in sentence "The joining of xenobiotic fragments with larger...." |
m The LinkTitles extension automatically added links to existing pages (<a rel="nofollow" class="external free" href="https://github.com/bovender/LinkTitles">https://github.com/bovender/LinkTitles</a>). |
||
| Line 15: | Line 15: | ||
The water soluble and reactive compounds formed from transformation are then conjugated to endogenous modules such as sugars or peptides. The joining of xenobiotic fragments with larger endogenous compounds decreases the toxicity of the xenobiotic while also making it easier to shuttle them around the cell during the last phase of compartmentalization. Glycosyltransferases are a large family of enzymes that catalyze the joining of nucleotide-diphosphate-sugars (usually in the form of Uridine diphosphate glucose or UDP-glucose) to xenobiotic compounds (6). The glycosylation of these compounds results in a higher stability of the xenobiotic fragments as well as trapping them within the vacuole and preventing them from reacting further to form harmful substances (7). Glutathione S-transferases (GSTs) is another class of enzymes that attach to xenobiotics at a tripeptide glutathione region and facilitate transfer to the vacuole or cell wall (8). | The water soluble and reactive compounds formed from transformation are then conjugated to endogenous modules such as sugars or peptides. The joining of xenobiotic fragments with larger endogenous compounds decreases the toxicity of the xenobiotic while also making it easier to shuttle them around the cell during the last phase of compartmentalization. Glycosyltransferases are a large family of enzymes that catalyze the joining of nucleotide-diphosphate-sugars (usually in the form of Uridine diphosphate glucose or UDP-glucose) to xenobiotic compounds (6). The glycosylation of these compounds results in a higher stability of the xenobiotic fragments as well as trapping them within the vacuole and preventing them from reacting further to form harmful substances (7). Glutathione S-transferases (GSTs) is another class of enzymes that attach to xenobiotics at a tripeptide glutathione region and facilitate transfer to the vacuole or cell wall (8). | ||
The final phase sees the xenobiotics bound to larger macromolecules sequestered to specific locations within plant cells, namely the vacuole or cell wall. Most xenobiotics are incorporated into the cell wall after being bound to lignin molecules while enzymes called ATP-binding cassette (ABC) transporters facilitate xenobiotic transfer to the vacuole (9). | The final phase sees the xenobiotics bound to larger macromolecules sequestered to specific locations within plant cells, namely the vacuole or cell wall. Most xenobiotics are incorporated into the cell wall after being bound to [[lignin]] molecules while enzymes called ATP-binding cassette (ABC) transporters facilitate xenobiotic transfer to the vacuole (9). | ||
== Rhizodegradation == | == Rhizodegradation == | ||
Latest revision as of 13:03, 6 May 2022
Definition
Phytoremediation is a process that uses vascular plants as a means of extracting inorganic and organic contaminants from soils (1). The strategies used in phytoremediation can be grouped into physical, chemical and biological methods for mitigating the effect subsurface pollutants have on the soil and groundwater.
Phytodegradation
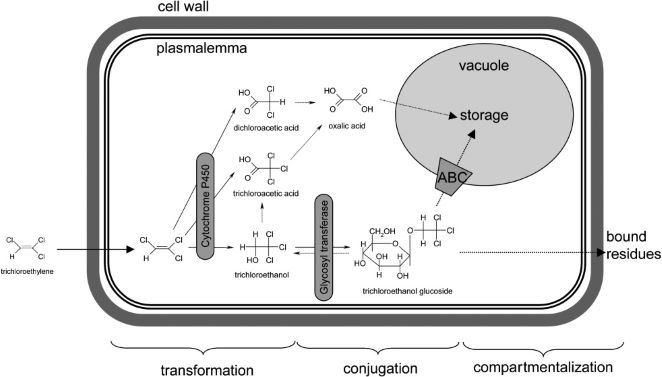
Phytodegradation utilizes the metabolic capability of plants in breaking down soil contaminants. The term “green liver” has been used to describe this process as the plant metabolizes xenobiotic compounds in an analogous way to that of the mammalian liver (2). In plants, the xenobiotic metabolism occurs over three phases; transformation, conjugation, and excretion. The result of these phases is detoxification of the xenobiotic as well as making them more inert and stored away from other cellular processes.
The first phase involves the chemical modification of the xenobiotic compound by either oxidation, reduction, or hydrolysis. This causes the xenobiotic to become more water soluble and thus be more biochemically reactive within the plant (3). Plants utilize many different enzymes to alter these compounds. Cytochrome P450 family enzymes act as mono-oxygenases towards a broad range of substrates and convert hydrophobic compounds into those which are more soluble in water (4). Carboxylesterases (CXEs) can convert carboxyl esters into carboxylic acids via hydrolysis which can go on to react with other molecules in the next phase (5).
The water soluble and reactive compounds formed from transformation are then conjugated to endogenous modules such as sugars or peptides. The joining of xenobiotic fragments with larger endogenous compounds decreases the toxicity of the xenobiotic while also making it easier to shuttle them around the cell during the last phase of compartmentalization. Glycosyltransferases are a large family of enzymes that catalyze the joining of nucleotide-diphosphate-sugars (usually in the form of Uridine diphosphate glucose or UDP-glucose) to xenobiotic compounds (6). The glycosylation of these compounds results in a higher stability of the xenobiotic fragments as well as trapping them within the vacuole and preventing them from reacting further to form harmful substances (7). Glutathione S-transferases (GSTs) is another class of enzymes that attach to xenobiotics at a tripeptide glutathione region and facilitate transfer to the vacuole or cell wall (8).
The final phase sees the xenobiotics bound to larger macromolecules sequestered to specific locations within plant cells, namely the vacuole or cell wall. Most xenobiotics are incorporated into the cell wall after being bound to lignin molecules while enzymes called ATP-binding cassette (ABC) transporters facilitate xenobiotic transfer to the vacuole (9).
Rhizodegradation
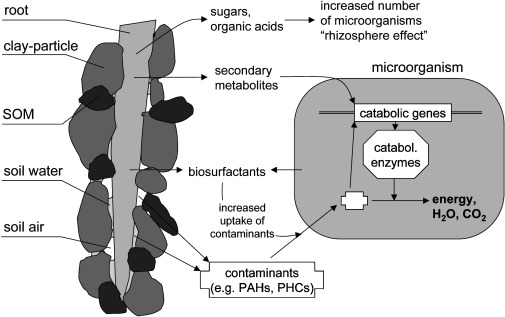
Rhizodegradation results from the establishment a vast variety of soil microorganisms in the region surrounds the roots known as the rhizosphere. This region serves as a favorable habitat and in turn increases the degradation of pollutants by the soil microorganisms themselves (1). Plants can lead to the induction of catabolic genes in soil microbes as well as population shifts through the release of root exudates (10). Sugars, amino acids, and organic acids are commonly released into the rhizosphere where they are readily used by microbes to grow. Along with these compounds’ plants release secondary metabolites for soil microbes, namely isoprenoids, alkaloids, and flavonoids. Normally soil microbes would not necessarily switch to using xenobiotic compounds as a source of carbon as it would be a negative return for them. However, these secondary compounds from plants mimic that of xenobiotics and are easier to break down (11). This in turn causes microbes to synthesize catabolic proteins that break down the secondary metabolites as well as the xenobiotic contaminants in the soil.
References
- Reichenauer, Thomas G., and James J. Germida. “Phytoremediation of Organic Contaminants in Soil and Groundwater.” ChemSusChem, vol. 1, no. 8‐9, WILEY‐VCH Verlag, 2008, pp. 708–17, doi:10.1002/cssc.200800125.
- Sandermann H Jr. Plant metabolism of xenobiotics. Trends Biochem Sci. 1992 Feb;17(2):82-4. doi: 10.1016/0968-0004(92)90507-6. PMID: 1566333.
- Komives T, Gullner G. Phase I xenobiotic metabolic systems in plants. Z Naturforsch C J Biosci. 2005 Mar-Apr;60(3-4):179-85. PMID: 15948581. https://pubmed-ncbi-nlm-nih-gov.gate.lib.buffalo.edu/15948581.
- Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006 Jun 25;124(1):128-45. doi: 10.1016/j.jbiotec.2006.01.026. Epub 2006 Mar 3. PMID: 16516322. https://pubmed-ncbi-nlm-nih-gov.gate.lib.buffalo.edu/16516322.
- D. Werck‐Reichhart, A. Hehn, L. Didierjean, Trends Plant Sci. 2000, 5, 116–123. https://www-sciencedirect-com.gate.lib.buffalo.edu/science/article/abs/pii/S1360138500015673?via%3Dihub.
- Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000 Sep;5(9):380-6. doi: 10.1016/s1360-1385(00)01720-9. PMID: 10973093. https://pubmed-ncbi-nlm-nih-gov.gate.lib.buffalo.edu/10973093.
- Kahn RA, Bak S, Svendsen I, Halkier BA, Møller BL. Isolation and reconstitution of cytochrome P450ox and in vitro reconstitution of the entire biosynthetic pathway of the cyanogenic glucoside dhurrin from sorghum. Plant Physiol. 1997 Dec;115(4):1661-70. doi: 10.1104/pp.115.4.1661. PMID: 9414567; PMCID: PMC158632. https://pubmed-ncbi-nlm-nih-gov.gate.lib.buffalo.edu/9414567.
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000 May;5(5):193-8. doi: 10.1016/s1360-1385(00)01601-0. PMID: 10785664. https://pubmed-ncbi-nlm-nih-gov.gate.lib.buffalo.edu/10785664.
- Philip A. Rea. “MRP Subfamily ABC Transporters from Plants and Yeast.” Journal of Experimental Botany, vol. 50, no. 90001, OXFORD UNIVERSITY PRESS, 1999, pp. 895–913, doi:10.1093/jexbot/50.suppl_1.895. https://www-jstor-org.gate.lib.buffalo.edu/stable/23696197?sid=primo&seq=1#metadata_info_tab_contents.
- Fatima, Kaneez, et al. “Bacterial Rhizosphere and Endosphere Populations Associated with Grasses and Trees to Be Used for Phytoremediation of Crude Oil Contaminated Soil.” Bulletin of Environmental Contamination and Toxicology, vol. 94, no. 3, Springer US, 2015, pp. 314–20, doi:10.1007/s00128-015-1489-5. https://web-a-ebscohost-com.gate.lib.buffalo.edu/ehost/detail/detail?vid=0&sid=241754d3-ccb3-4acc-83b9-afafa5c82bcb%40sdc-v-sessmgr03&bdata=JnNpdGU9ZWhvc3QtbGl2ZSZzY29wZT1zaXRl#AN=101049230&db=eih.
- Singer AC, Crowley DE, Thompson IP. Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol. 2003 Mar;21(3):123-30. doi: 10.1016/S0167-7799(02)00041-0. PMID: 12628369. https://pubmed-ncbi-nlm-nih-gov.gate.lib.buffalo.edu/12628369.