Polymerase Chain Reaction (PCR): Difference between revisions
| (16 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
= | =Background= | ||
Polymerase chain reaction (PCR) was first created in 1983 by biochemist Kary Mullins [3]. This method was first created as a way to pinpoint certain strands of DNA and create synthetic copies of it in order to examine it better [1,2]. Before PCR studying DNA was more difficult as it was hard to isolate small strands of DNA and study them [2]. Kary Mullins would go on to win the 1993 Nobel Prize in Chemistry for the creation of PCR along with Michael Smith for his work on PCR [2]. Since winning this award in 1993 there have been only been four other years where the Nobel Prize in Chemistry was awarded for a method (1998,2002,2005,2020)[7]. | |||
=Definition= | |||
PCR is a method used to amplify a small amount of DNA in order to study it in detail[1]. RNA can also be extracted from samples and converted into complimentary DNA (cDNA) for PCR amplification [6]. Primers are used to identify the location of the DNA in the sample. Enzymes that have defined segments of DNA are utilized to recreate cDNA [6]. | |||
==PCR Components== | |||
In order to conduct PCR there are numerous components that are required. | |||
* DNA template or cDNA: These are regions that will be getting amplified [5] | |||
*Two primers: This determines the beginning and end of the DNA template or cDNA being amplified [5] | |||
*Taq polymerase: This copies the region being amplified [5] | |||
*Nucleotides: This is from the DNA polymerase for the new DNA [5] | |||
*Buffer: This provides a chemical environment for the DNA-Polymerase [5] | |||
==Primers== | ==Primers== | ||
PCR primers are single strands of | PCR primers are single strands of DNA used to identify the location of the DNA in the sample. This refers to a small set of nucleotides in DNA. The use of primers corresponds with the beginning and end of the DNA fragment to be amplified. They stick to the DNA template at the beginning and end, where the DNA-Polymerase binds and starts synthesis of new DNA strand [1,2,4,8]. For bacteria and [[archaebacteria]] primers that are ubiquitous to the 16s ribosomal RNA (rRNA) are used [1,2,4,8,9]. | ||
[[File:Screen Shot 2021-04-15 at 3.51.44 PM.png|thumb|right|Stages of PCR and the resultant amplification of DNA copies of the target region[2]]] | [[File:Screen Shot 2021-04-15 at 3.51.44 PM.png|thumb|right|Stages of PCR and the resultant amplification of DNA copies of the target region[2]]] | ||
==Stages of PCR== | ==Stages of PCR== | ||
'''Denaturing Stage:''' The denaturing stage is where the double-stranded DNA is heated up anywhere from 94-100<sup>oC</sup> to separate the strands which are held together by hydrogen bonds. This is often done for up to 5 minutes to ensure that both the template and the primer have completely seperated and are single strand only. Taq polymerase is also activated during this stage [5,6]. | |||
'''Annealing Stage:''' During the annealing stage the temperature is lowered to about 50<sup>oC</sup> below the specific primers melting temperature so the primers can attach themselves to themselves to single DNA strands to create double-stranded DNA. The temperature in this stage is critical as the wrong temperature can result in primers not binding to template DNA at all or binding at random [5,6] | |||
'''Extension Stage:''' Following the annealing stage comes extension. During this stage DNA polymerase (an enzyme) catalyzes the the synthesis of the new strands of DNA. With the annealed primer DNA polymerase adds complimentary nucleotides complimentary to the unpaired DNA strand [5,6]. | |||
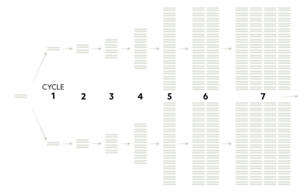
These steps are then repeated 20-40 to potentially create millions or billions amount of copied DNA of the target sequence. | |||
[[File:PCR cyclePM.png|thumb|left|Multiple completed cycles of PCR [7]]] | |||
==Uses== | ==Uses== | ||
PCR is helpful as it recreates small strands of DNA using either DNA or RNA. This is especially helpful in looking at genetic [[ecology]] studies as it allows a closer look at DNA [8]. It allows for the understanding of gene expression either spatially or temporally among [[organisms]] [1]. Bruce et al. (1992) used this method to study DNA sequences of native bacterial populations in [[soil]], [[sand]], and sediment [1]. Bacterial cultures is the traditional way to sample these, but it usually only accounts for a small amount of microbial biomass [1]. Bacterial cultures do not work well for slow-growing [[microorganisms]] such as mycobacteria and anaerobic bacteria [5]. PCR is able to be done rapidly and effectively making it a common practice across the science community. Pathogens are among samples that are able to be seen using PCR [1,6,9]. Today PCR is widely known for detecting pathogens such as Covid-19. | |||
==References== | ==References== | ||
1 | [1] Bruce, K.D., Hiorns, W.D., Hobman, J.L., Osborn, A.M., Strike, P., Ritchie, D.A., 1992. Amplification of DNA from native populations of [[soil]] bacteria by using the polymerase chain reaction. Applied and Environmental Microbiology 58, 3413–3416. https://doi.org/10.1128/AEM.58.10.3413-3416.1992 | ||
[2] Henson, J.M., French, R.C., n.d. THE POLYMERASE CHAIN REACTION AND PLANT DISEASE DIAGNOSIS 30. | |||
[3] Kossakovski, F., n.d. The eccentric scientist behind the ‘gold standard’ COVID-19 test [WWW Document]. National Geographic. URL https://www.nationalgeographic.com/science/article/the-eccentric-scientist-behind-the-gold-standard-covid-19-pcr-test | |||
[4] Picard, C., Ponsonnet, C., Paget, E., Nesme, X., Simonet, P., 1992. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Applied and Environmental Microbiology 58, 2717–2722. https://doi.org/10.1128/AEM.58.9.2717-2722.1992 | |||
[5] Rahman, M.T., Uddin, M.S., Sultana, R., Moue, A., Setu, M., 2013. Polymerase Chain Reaction (PCR): A Short Review. Anwer Khan Mod Med Coll J 4, 30–36. https://doi.org/10.3329/akmmcj.v4i1.13682 | |||
[6] Schochetman, G., Ou, C.-Y., 2021. Polymerase Chain Reaction 5 | |||
[7] The Nobel Prize in Chemistry [WWW Document], n.d. . nobelprize. URL https://www.nobelprize.org/prizes/chemistry/ | |||
[8] Tsai, Y.L., Olson, B.H., 1992. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Applied and Environmental Microbiology 58, 754–757. https://doi.org/10.1128/AEM.58.2.754-757.1992 | |||
[9] WILSONl, K.H., Blitchington, R.B., Greene, R.C., 1990. Amplification of Bacterial 16S Ribosomal DNA with Polymerase Chain Reaction. J. CLIN. MICROBIOL. 28, 5. | |||
Latest revision as of 10:39, 7 May 2021
Background
Polymerase chain reaction (PCR) was first created in 1983 by biochemist Kary Mullins [3]. This method was first created as a way to pinpoint certain strands of DNA and create synthetic copies of it in order to examine it better [1,2]. Before PCR studying DNA was more difficult as it was hard to isolate small strands of DNA and study them [2]. Kary Mullins would go on to win the 1993 Nobel Prize in Chemistry for the creation of PCR along with Michael Smith for his work on PCR [2]. Since winning this award in 1993 there have been only been four other years where the Nobel Prize in Chemistry was awarded for a method (1998,2002,2005,2020)[7].
Definition
PCR is a method used to amplify a small amount of DNA in order to study it in detail[1]. RNA can also be extracted from samples and converted into complimentary DNA (cDNA) for PCR amplification [6]. Primers are used to identify the location of the DNA in the sample. Enzymes that have defined segments of DNA are utilized to recreate cDNA [6].
PCR Components
In order to conduct PCR there are numerous components that are required.
- DNA template or cDNA: These are regions that will be getting amplified [5]
- Two primers: This determines the beginning and end of the DNA template or cDNA being amplified [5]
- Taq polymerase: This copies the region being amplified [5]
- Nucleotides: This is from the DNA polymerase for the new DNA [5]
- Buffer: This provides a chemical environment for the DNA-Polymerase [5]
Primers
PCR primers are single strands of DNA used to identify the location of the DNA in the sample. This refers to a small set of nucleotides in DNA. The use of primers corresponds with the beginning and end of the DNA fragment to be amplified. They stick to the DNA template at the beginning and end, where the DNA-Polymerase binds and starts synthesis of new DNA strand [1,2,4,8]. For bacteria and archaebacteria primers that are ubiquitous to the 16s ribosomal RNA (rRNA) are used [1,2,4,8,9].

Stages of PCR
Denaturing Stage: The denaturing stage is where the double-stranded DNA is heated up anywhere from 94-100oC to separate the strands which are held together by hydrogen bonds. This is often done for up to 5 minutes to ensure that both the template and the primer have completely seperated and are single strand only. Taq polymerase is also activated during this stage [5,6].
Annealing Stage: During the annealing stage the temperature is lowered to about 50oC below the specific primers melting temperature so the primers can attach themselves to themselves to single DNA strands to create double-stranded DNA. The temperature in this stage is critical as the wrong temperature can result in primers not binding to template DNA at all or binding at random [5,6]
Extension Stage: Following the annealing stage comes extension. During this stage DNA polymerase (an enzyme) catalyzes the the synthesis of the new strands of DNA. With the annealed primer DNA polymerase adds complimentary nucleotides complimentary to the unpaired DNA strand [5,6].
These steps are then repeated 20-40 to potentially create millions or billions amount of copied DNA of the target sequence.
Uses
PCR is helpful as it recreates small strands of DNA using either DNA or RNA. This is especially helpful in looking at genetic ecology studies as it allows a closer look at DNA [8]. It allows for the understanding of gene expression either spatially or temporally among organisms [1]. Bruce et al. (1992) used this method to study DNA sequences of native bacterial populations in soil, sand, and sediment [1]. Bacterial cultures is the traditional way to sample these, but it usually only accounts for a small amount of microbial biomass [1]. Bacterial cultures do not work well for slow-growing microorganisms such as mycobacteria and anaerobic bacteria [5]. PCR is able to be done rapidly and effectively making it a common practice across the science community. Pathogens are among samples that are able to be seen using PCR [1,6,9]. Today PCR is widely known for detecting pathogens such as Covid-19.
References
[1] Bruce, K.D., Hiorns, W.D., Hobman, J.L., Osborn, A.M., Strike, P., Ritchie, D.A., 1992. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Applied and Environmental Microbiology 58, 3413–3416. https://doi.org/10.1128/AEM.58.10.3413-3416.1992
[2] Henson, J.M., French, R.C., n.d. THE POLYMERASE CHAIN REACTION AND PLANT DISEASE DIAGNOSIS 30.
[3] Kossakovski, F., n.d. The eccentric scientist behind the ‘gold standard’ COVID-19 test [WWW Document]. National Geographic. URL https://www.nationalgeographic.com/science/article/the-eccentric-scientist-behind-the-gold-standard-covid-19-pcr-test
[4] Picard, C., Ponsonnet, C., Paget, E., Nesme, X., Simonet, P., 1992. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Applied and Environmental Microbiology 58, 2717–2722. https://doi.org/10.1128/AEM.58.9.2717-2722.1992
[5] Rahman, M.T., Uddin, M.S., Sultana, R., Moue, A., Setu, M., 2013. Polymerase Chain Reaction (PCR): A Short Review. Anwer Khan Mod Med Coll J 4, 30–36. https://doi.org/10.3329/akmmcj.v4i1.13682
[6] Schochetman, G., Ou, C.-Y., 2021. Polymerase Chain Reaction 5
[7] The Nobel Prize in Chemistry [WWW Document], n.d. . nobelprize. URL https://www.nobelprize.org/prizes/chemistry/
[8] Tsai, Y.L., Olson, B.H., 1992. Detection of low numbers of bacterial cells in soils and sediments by polymerase chain reaction. Applied and Environmental Microbiology 58, 754–757. https://doi.org/10.1128/AEM.58.2.754-757.1992
[9] WILSONl, K.H., Blitchington, R.B., Greene, R.C., 1990. Amplification of Bacterial 16S Ribosomal DNA with Polymerase Chain Reaction. J. CLIN. MICROBIOL. 28, 5.